About the DISEASE OUTBREAKS DATA project
The DISEASE OUTBREAKS DATA project arose from the need for open, reliable information on pandemic- and epidemic-prone disease outbreaks, offering broad coverage of diseases, time periods, and geography, and ensuring statistical soundness for research purposes.
The dataset is the result of a collaborative effort by a team of researchers from the University of Göttingen, the University of Groningen, and the University of Bordeaux. The project was made possible through financial support from the ENLIGHT network, the German Academic Exchange Service (DAAD), and the Federal Ministry of Education and Research (BMBF) in Germany.
In the first version of the dataset, a total of 2227 outbreaks of 70 different infectious diseases were found, occurring in a total of 233 countries and territories from January 1996 until March 2022. These findings are published in Springer Nature’s Scientific Data. Read the paper by clicking here! Additionally, the data, metadata, and the code to replicate the first version of this dataset are publicly available on Figshare. You can download them by clicking here!.
The unit of analysis in the database is an outbreak, defined as the occurrence of at least one case of a specific disease in a country -or territory- during a particular year. Therefore, a country -or territory- cannot have more than one outbreak of the same disease in the same year, although it may experience outbreaks of different diseases within the same year. A country can only have multiple outbreaks of the same disease if they occur in different years.
The last version of the dataset contains information on 3451 outbreaks:
Temporal coverage: 01/01/1996 - 26/01/2026
Geographic coverage: 236
Number of diseases included: 91
Methodology
Source
The sources for the DISEASE OUTBREAKS DATA project are the Disease Outbreak News (DONs) and the Coronavirus Dashboard produced by the World Health Organization (WHO). This information is issued under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Intergovernmental Organization (CC BY-NC-SA 3.0 IGO) license, which allows users to freely copy, reproduce, reprint, distribute, translate, and adapt WHO materials for non-commercial purposes.
The information from the DONs includes all reports on confirmed acute public health events or potential events of concern that have occurred since 1996. Specifically, DONs include events of:
- Unknown cause but with significant or potential health concern that may affect international travel or trade.
- Known cause with demonstrated ability to produce a serious public health impact and spread internationally.
- High public concern that could potentially disrupt required public health interventions or international travel and trade.
The Coronavirus Dashboard presents information reported by official public health authorities from countries and territories worldwide.
Data collection and integration processes
The following figure provides a schematic overview of the data collection and integration processes used in the disease outbreaks data project.
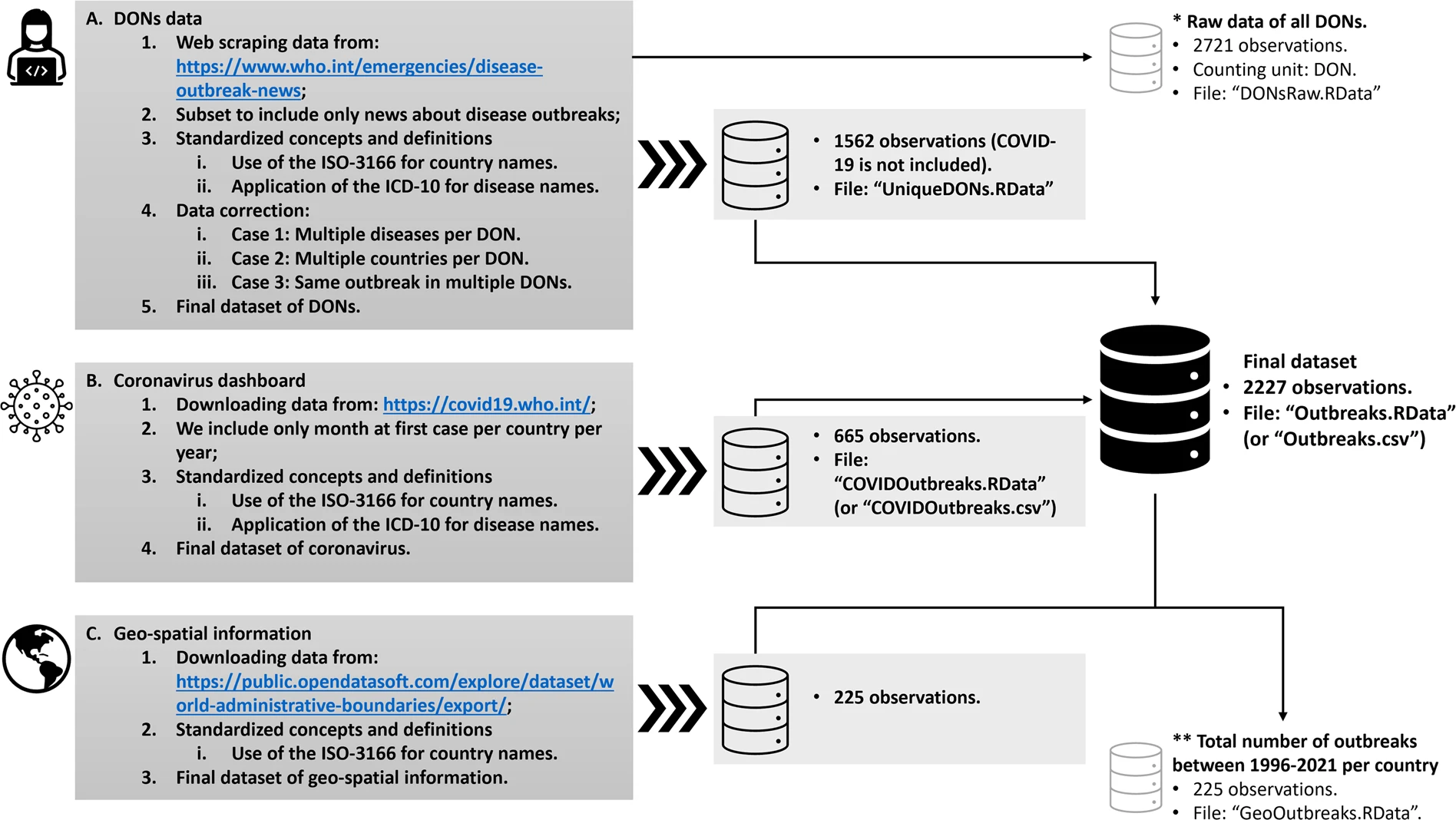
Source: Torres Munguía, Badarau, Díaz Pavez, Martínez-Zarzoso & Wacker. A global dataset of pandemic- and epidemic-prone disease outbreaks. Sci Data 9, 683 (2022). https://doi.org/10.1038/s41597-022-01797-2
In stage (A), DONs are collected from the WHO website. This process was automated using an R script to extract the information from the DONs. The earliest DON records a cholera outbreak reported on 22 January 1996 in Cabo Verde, Côte d’Ivoire, the Islamic Republic of Iran, Iraq, and Senegal.
To ensure standardized concepts and definitions, official short country names in English, according to ISO-3166-23 and International Statistical Classification of Diseases and Related Health Problems 10th Revision, are used.
Three recording issues need to be tackled at stage (A):
Some DONs report multiple diseases.
Some DONs report disease outbreaks occurring in more than one country.
Some DONs register the same outbreak multiple times due to situation updates.
To resolve these issues at stage (A):
For DONs reporting more than one disease (for instance, DON0065 on influenza and malaria in Ghana, or DON1094 on chikungunya and dengue in the southwest Indian Ocean) and/or reporting more than one country (e.g., DON1540 about an outbreak of polio in Angola and the Democratic Republic of the Congo, or DON0617 on a meningococcal disease outbreak in the Great Lakes area) the DON is replicated for each diseases (or country). For instance, DON0617 informs of an outbreak that occurred in Burundi, Rwanda, and Tanzania (Great Lakes area). Therefore, this DON was registered three times, one for each country.
To avoid multiplicity issues, we deleted all DONs that reported the same disease in the same country more than once in a calendar year. Variants or mutations of viruses, such as avian influenza A(H1N1), A(H1N2), A(H5N1), A(H3N2), etc., were considered the same disease, i.e., influenza due to identified zoonotic or pandemic influenza virus. This ensured only one observation per disease, country, and year.
In stage (B), given that outbreaks related to COVID-19 are not included in the DONs, this information is extracted from the Coronavirus Dashboard. Specifically, we dichotomized the data on cases per country per year, assigning a value of one if a country had at least one reported case of Coronavirus, and zero otherwise. For standardization, we followed the same approach as in stage (A), using the official short country names in English according to ISO-3166-23 and ICD-10.
In stage (C), the geographic information from the World Administrative Boundaries - Countries and Territories dataset by Société OPENDATASOFT (available at this link) is merged with the resulting data from stages (A) and (B).